Soichiro, Our Head of Technology will have a poster session at Enzyme Engineering XXVI in Texas, US from 22th May to 27th May.
Come learn our database and enzyme discovery pipeline!
Title: Efficient enzyme discovery from complex environmental microbiota using microbial single-cell sequencing
Presentor: Soichiro Tsuda, Head of Technology, bitBiome, Inc.
Session date: 23-24 May, 2022
Abstract:
Environmental microbiota is a vast untapped resource for novel biocatalysis enzymes. Shotgun metagenomic sequencing has been an effective tool to discover such enzymes from complex microbiotas, yet many challenges still remain especially for sequencing of diverse soil microbial genomes. We developed a microbial single-cell sequencing method, called bit-MAP®, where individual bacteria are isolated and their genomes are amplified in microfluidic gel capsules.
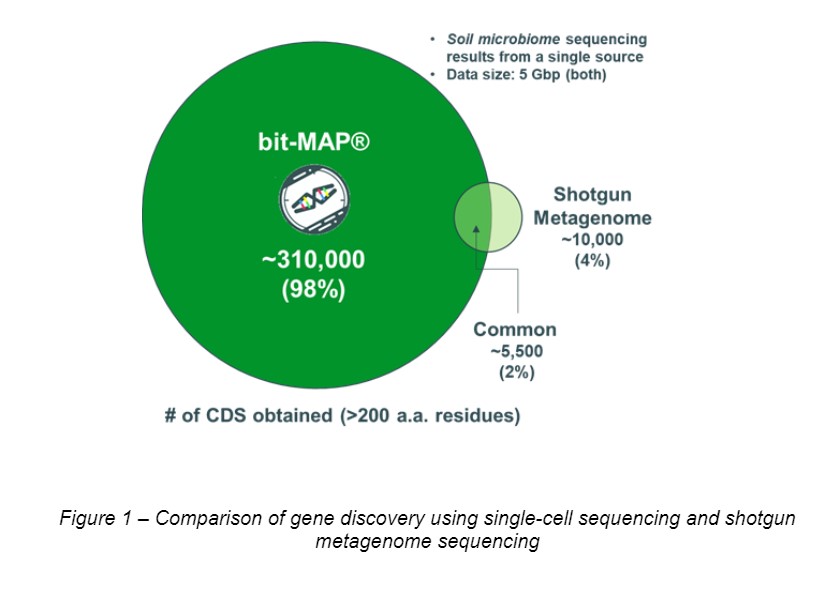
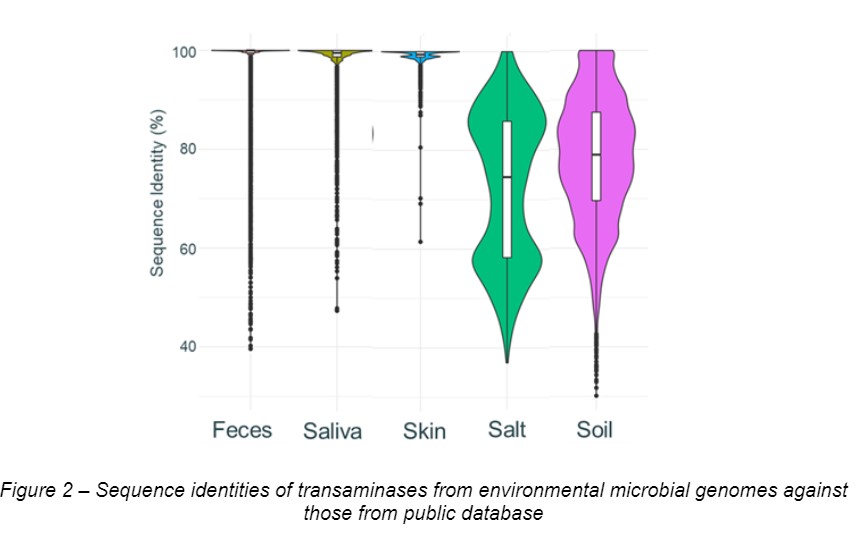
We found this method could recover microbial genes from complex environmental microbiota, more than 30 times efficient compared to the shotgun metagenomic sequencing (See Figure 1 below). Using this method, we have been constructing a massive microbial gene database containing over 300 million genes, which will be expanded up to 800 million by the end of the year. Enzymes from our database (especially from environmental microbiota) often show low sequence identity with enzymes from public database (Figure 2). At the conference, we will show our initial results for characterizing novel transaminases mined from our microbial gene database.
About bitBiome, Inc.
bitBiome will construct the world’s No.1 microbial genome database, in terms of quantity, quality, variety, and rarity, and will “renovate” the industrial structure through the application of microbial genes.