bitBiome is pleased to announce the official launch of its “Enzyme Discovery Service” utilizing a world’s largest massive microbial genome database (MMGDB) and our proprietary bioinformatics technology. The MMGDB has been constructed using bitBiome’s proprietary microbial single-cell genome sequencing technology, bit-MAP®, and thus contains a wide range of unculturable microbial genomes that were difficult to sequence with existing sequencing technologies. We also offer “in silico DOE Service” using in silico mutation analysis through sequence variant analysis and 3D structural modeling.
Our enzyme discovery service and enzyme modification support service can be offered separately or in combination. If you are interested, please do not hesitate to contact us service@bitbiome.co.jp
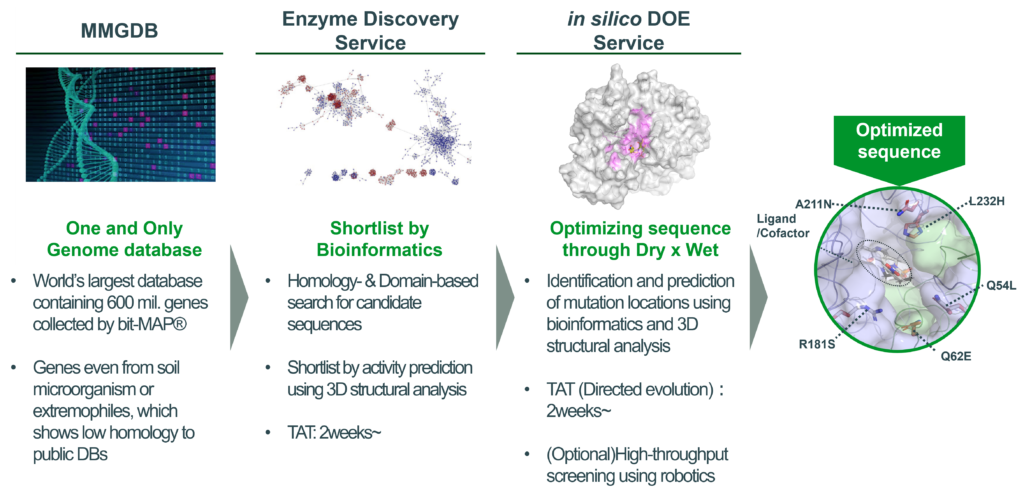
■MMGDB: A world’s largest unique microbial genome database
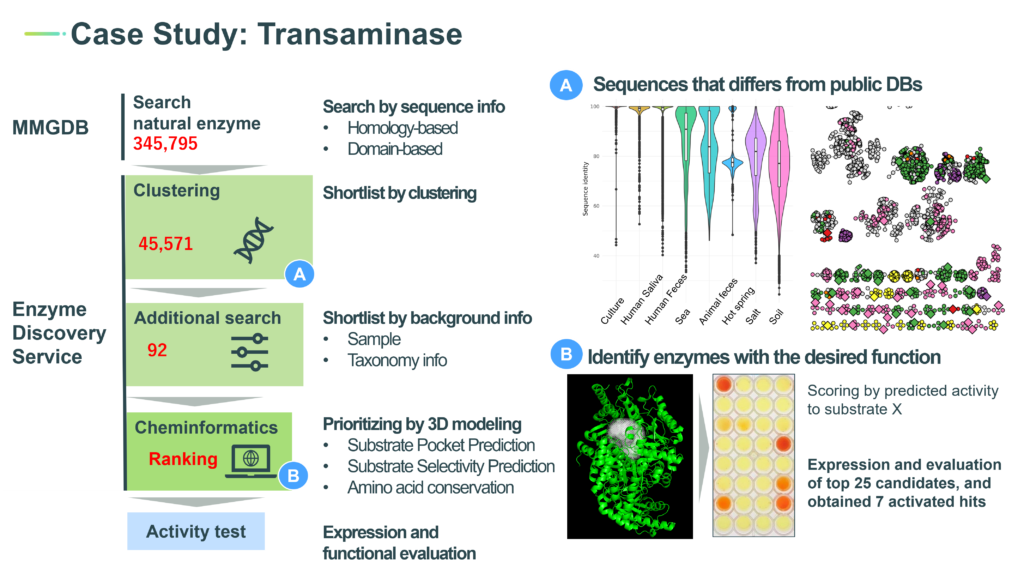
bitBiome uses its core technology, bit-MAP®, a microbial single-cell genome analysis technology, to analyze the human microbiome as well as all environmental microbes living in soil, ocean, hot springs or etc. to create the MMGDB. This DB currently contains over 600 million genes (as of July 2022), which is the world’s largest database in private companies (based on publicly available data).
The MMGDB features a number of unique genes derived from soil microorganisms and other difficult-to-culture microorganisms. We confirmed that they are largely different from genes in public databases in terms of sequence identities, and hence ready to be utilized for a wide range of synbio and biocatalysis applications.
■Enzyme Discovery Service: Search optimized enzymes using bioinformatics and cheminformatics
bitBiome provides enzyme discovery services tailored for client’s requests to search optimal sequences from the MMGDB utilizing its proprietary bioinformatics and 3D structural modeling technologies.
It only takes three weeks from searching the target enzyme to returning the result.
■in silico DOE service:
bitBiome can also optimize enzymes by in silico (dry) enzyme analysis. Mutation sites for directed evolution of a target gene can be predicted based on the comparative gene analysis and 3D structural modeling. Optionally, activities and characteristics can be improved in a short period of time through efficient wet-lab experiments using automated liquid handling robotics.