Achieve unparalleled value creation by unlocking the potential of microbes
Microbes are the earth’s richest, yet most underutilized resources. Their genetic functions are critical to develop new medicines and climate change solutions.
With our Gene Encyclopedia from Microbes (bit-GEM) built by bit-MAP® single-cell genomics we help clients address their big issues by uncovering novel microbial functions and improving them .
Enzyme Discovery & Engineering Service
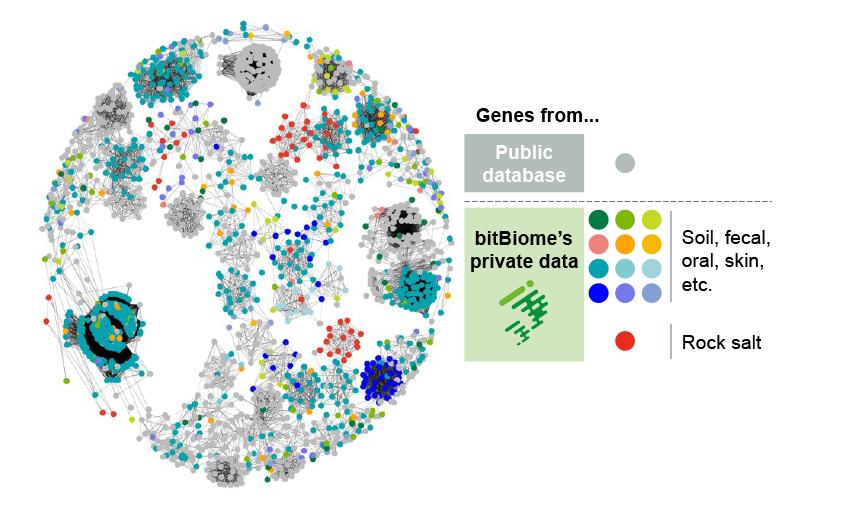
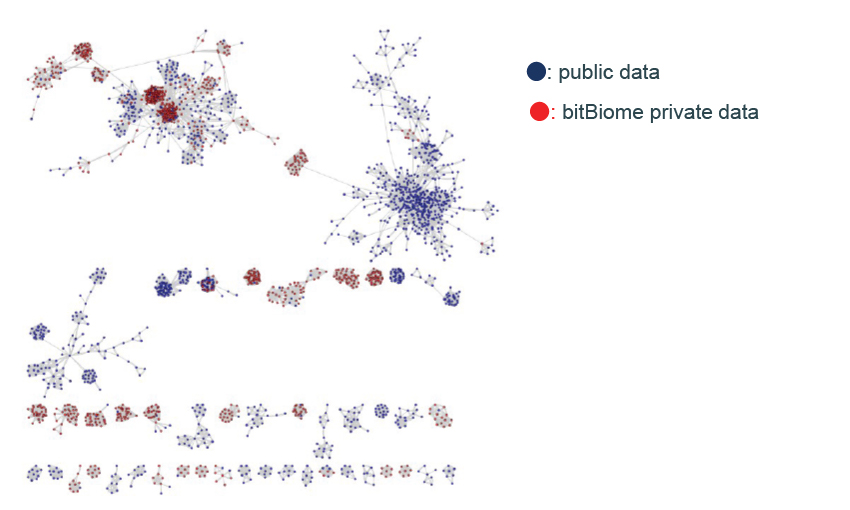
Our proprietary Gene Encyclopedia from Microbes (bit-GEM) contains over 800 million microbe genes including unculturables and extremophiles. With this bit-GEM, highly effective sequencing platform and a team of bioinformatics experts, we help you identify and optimize candidate genes.
Gene discovery at
bit-GEM

The one and only
genome database
- Over 800 million genes from single-cell bacteria genomes
- Rich genetic diversity including unculturable bacteria and extremophile
- Can discover unique enzymes with low homology to public DBs

Bio-informatics
- Comprehensive bioinformatics support from
- Shortlist candidate enzymes through bioinformatic and 3D structural analyses
- In silico design-ofexperiments support for directed evolution

Speed
- Significantly faster than standard methods using public database
- Delivery timing(*)
- Bioinformatics : ~1
month - Wet evaluation (**) report:
~6 months

Flexibility
- Tailor-made support (dry & wet) based on your needs
- Consult on the choice of candidate enzymes ahead of a full contract
- Flexible forms of contracts
(*) Best effort basis
(**)After specifying evaluation scope and methods
SynBio Applications

Plastics Degrading Enzyme
bit-GEM contains a huge number of unique genes useful for environmental problems. In particular, bitBiome with bit-QED has been developing an enzyme that can degrade polyethylene terephthalate (PET), which is currently the focus of worldwide interest.
1. 35,000 candidates found in bit-GEM
2. in silico prediction model chose several candidates from 35,000 candidates
3. Wet Evaluation using high-throughput screening system
4. Directed Evolution improved their activity
Genes for Synthetic Biology
We boost your in Design-Build-Test-Learn cycle by turbocharging Design phase, shortlisting candidate genes through bit-GEM search and bioinformatics.
Microbiome Sequencing Service
bit-MAP®sequencing service
We’re providing novel Single Cell Genomics analysis service at the competitive cost, under USD45 per genome (SAG) at 384-well plate, and the deliverable report contains rich contents (e.g. 1. Raw Sequence Data 2. Quality of Draft Genome 3. Phylogenetic analysis 4. Taxonomy Identification of each draft genome). (Demo Data. Gut Microbiome)
bit-MAP® is applicable to a wide range of microbial samples, including human microbiome (e.g., gut, skin and oral) and environmental bacteria (e.g., soil and marine).
We offer a comprehensive support for industrial researchers or academic researchers to unlock the potential of microbes through bit-MAP.
Please contact us for details.